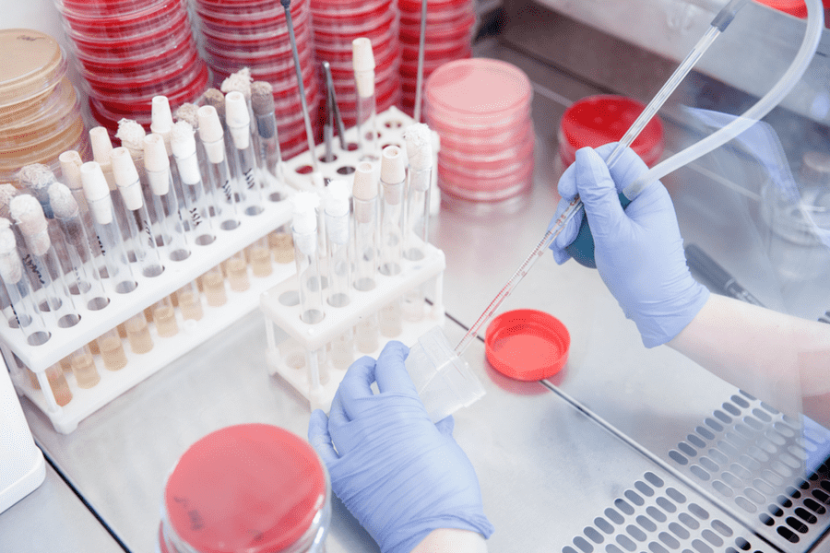
Finding a Needle in Haystack: Why Sample Concentration is Important Prior to Rapid Detection
By Dr. Matthew D. Moore, Sloane Stoufer, Anand Soorneedi
One of the most exciting areas of research in the realm of food safety has been the development of many rapid, culture-free detection techniques and technologies. There are now numerous types of detection technologies for pathogens in foods and food processing environments that potentially offer portability, rapidity, and analytical sensitivity. With increasing pressure on the food supply and resulting costs in food production and service environments, rapid detection technologies remain of crucial importance.
Dangerous Needles in Tasty Haystacks
With increasing pressure on the food supply and resulting costs in food production and service environments, rapid detection technologies remain of crucial importance.
Despite reports of promising pathogen detection technologies, the need for equally promising and effective upstream sample processing and concentration techniques for pathogens is often overlooked. This despite being crucial for rapid, culture-free detection of contaminants in foods and the environment. Unlike in many clinical applications, where the level of an infectious agent is comparably high, pathogens in food and environmental samples often occur in quite low levels and are unevenly distributed, making it difficult to obtain a representative sample. In the case of bacterial and fungal pathogens, this can often be mitigated by the inclusion of an enrichment (culture) steps prior to the utilization of a detection technology. However, these enrichment steps can add hours to days to the detection process. Further, such steps are not realistically applicable for viruses, toxins, and small molecule contaminants that may be present in foods or the environment. Thus, there is a need for other methods to concentrate the target pathogen/contaminant. Further, many rapid detection technologies are limited in terms of the volume of sample that can be analyzed (often less than 1 mL), even though the recommended size for representative food samples is typically 25 grams or more. Essentially, it is like trying to capture and concentrate finding a needle in a haystack.
One great example of a pathogen that poses such challenges to routine detection in foods and the environment is human norovirus, the leading cause of foodborne illness. Norovirus is estimated to require only a handful of particles to cause illness in individuals and can persist in the environment for weeks. Further, this virus is highly transmissible and can be rapidly spread, making portable, rapid detection important for reducing illness. There are two general types of methods by which this virus is concentrated, often used together sequentially: nonspecific and specific concentration methods.
Comparing Concentration Methods
Nonspecific concentration methods often exploit general physiochemical properties of a specific target. For example, filtration-based techniques exploit the fact that noroviruses are all similar in size (~38 nm in diameter), allowing larger particles to be separated from the virus. Another characteristic of noroviruses is that they are nonenveloped, meaning they have a protein capsid enclosing an RNA genome with no lipid envelope and therefore behave like proteins in an aqueous suspension. Thus, polyethylene glycol (PEG) is often utilized, as it causes most of the proteins in a sample to crash out of suspension. However, food and environmental samples have a lot of other compounds that behave similarly and are often co-concentrated with the target. Unfortunately, this other food and environmental “junk” can inhibit many downstream rapid detection techniques.
Thus, specific concentration methods are often required. Many of these specific concentration techniques utilize a ligand (like an antibody) that only binds and captures the target pathogen/contaminant. Immunomagnetic separation is an example in which antibodies specific for a pathogen are tethered onto magnetic beads; the antibodies bind the pathogen, and then are captured by a magnet placed on the outside of the tube containing the sample. The beads stay on the magnet while the rest of the sample can be removed and further washed to remove the aforementioned “junk,” leaving just a small sample of mostly concentrated target pathogen. However, these techniques currently cannot be realistically scaled up to volumes required for routine food and environmental testing in most cases, and have limitations related to their cost and/or throughput. This is especially true for noroviruses, and likely one of the major factors in the lack of routine testing for norovirus in food and environmental samples.
However, this area of sample pre-processing and concentration has been of increasing interest recently. Exciting work on novel sample processing technologies is ongoing, including some of the work the lab at Amherst is conducting. For example, our lab is currently developing a technique for norovirus concentration similar to the immunomagnetic separation technique mentioned above; however, we have replaced antibody-coated beads with bacteria that present ligands that specifically bind norovirus. Thus, one can simply culture as much bacteria as needed, pellet the bacteria, and use it in place of magnetic beads. In addition to being much larger and having higher surface area than most magnetic beads (more viruses per particle than a bead), it is much cheaper and easier to simply grow as much bacteria as needed, essentially allowing more room for scale-up to larger samples. Additionally, our lab is investigating the use of magnetic ionic liquids, an emerging reagent being investigated in the sample preparation space that has the potential to obviate the need for centrifugation or complex instrumentation prior to detection.
Thus, considering the many exciting developments in rapid detection technologies in the past couple decades, there is a growing need for upstream sample processing and concentration methods in order to fully utilize these technologies. The development of such sample processing techniques is continuing to receive attention from researchers with many exciting projects being investigated and in the next decade breakthroughs in this area may enable us to unlock the full potential of many developed rapid detection technologies.
Acknowledgements/Special PS: The bacterial concentration work described is supported by the USDA National Institute of Food and Agriculture, Agricultural and Food Research Initiative Competitive Program, grant number: 2019-67017-29588.
About the Author:
Dr. Matthew D. Moore is an Assistant Professor and director of the Applied and Environmental Virology Lab in the Department of Food Science at the University of Massachusetts, Amherst. The Applied and Environmental Virology Lab conducts research on numerous aspects of applied microbiology/virology, with a variety of research topics. This article was written in collaboration with Sloane Stoufer and Anand Soorneedi from the Department of Science, University of Massachusetts, Amherst MA. The Applied and Environmental Virology Lab conducts research on numerous aspects of applied microbiology/virology, with a variety of research topics, such as: understanding the gut microbiota and its role in eukaryotic viral pathogenesis; development of novel pathogen/contaminant concentration and detection technologies; understanding the mechanisms of viral environmental persistence and transmission; and developing novel inactivation formulations and materials for inactivation and/or removal of pathogens/contaminants in foods and the environment. The lab is always actively seeking research partners from industry, academia, and government; anyone interested is encouraged to email Dr. Moore at: [email protected].

-
 FeaturedRisk management
The Cost of a Breach: What a Cyberattack Could Mean for Food Safety Recalls
FeaturedRisk management
The Cost of a Breach: What a Cyberattack Could Mean for Food Safety Recalls
-
 FeaturedRisk management
Securing the Food Chain: How ISO/IEC 27001 Strengthens Cybersecurity
FeaturedRisk management
Securing the Food Chain: How ISO/IEC 27001 Strengthens Cybersecurity
-
 FeaturedRisk management
Revolutionizing Food Safety Training: Breaking Out of the “Check-the-Box” Mentality
FeaturedRisk management
Revolutionizing Food Safety Training: Breaking Out of the “Check-the-Box” Mentality
-
 GFSI Standards
GFSI 2025: Building Trust, Tech-Forward Solutions, and Global Unity in Food Safety
GFSI Standards
GFSI 2025: Building Trust, Tech-Forward Solutions, and Global Unity in Food Safety
-
 FeaturedFood Safety
Integrated Pest Management: Strategies to Protect Your Brand’s Reputation
FeaturedFood Safety
Integrated Pest Management: Strategies to Protect Your Brand’s Reputation
-
 FeaturedFood Safety Culture & Training
No Open Door Policy: Challenges That Impact Pest Control in Food Processing Plants
FeaturedFood Safety Culture & Training
No Open Door Policy: Challenges That Impact Pest Control in Food Processing Plants